Real time genomic surveillance of Ebola outbreak 2014-2015
05 Jun 2015The current Ebola outbreak in West Africa is the largest ever recorded, with over 26,500 cases reported resulting in an estimated 11,000 deaths. Yet genomic surveillance of this outbreak has been patchy, hampered by understandable but vexing logistical, social, political and technical obstacles in securing and transporting samples for processing.
We wanted to help address the gaps in our knowledge of viral evolution and to generate data for epidemiological use. So, in April, Josh Quick from my group went to Conakry, Guinea to establish proof-of-principle for portable nanopore sequencing. This was the most practical way we could rapidly establish a local sequencing lab in order to generate real-time information.
His travels have been documented in several recent news articles. For background I would recommend reading Erika Hayden’s report over at Nature News, the BMC On Biology blog and this recent GenomeWeb article (registration free for academic subscribers).
In the two weeks he was there, he sequenced 14 genomes when based at Donka Hospital in Conakry. However, the surveillance sequencing has continued, thanks to the hard work of Sophie Duraffour in Coyah under the auspcies of the European Mobile Laboratory project. Sophie has been working around the clock in the laboratory generating the real-time genome data, uploading it to Birmingham for analysis and then distributing it to WHO central coordination. We have had early feedback that the data has been extremely useful for the epidemiologists on the ground.
As is often the case in outbreaks, genomic data production and sharing has been patchy and uncoordinated. However, a new exciting deveopment is under way to try and address this. Andrew Rambaut, author of essential phylogenetics software such as BEAST and FigTree and viral genome maven, has taken on a kind of unofficial role of coordinating genome sequence data, which is distributed through his website and forum Virological.org.
His personal database of Ebola genomes sits at nearly 1000 sequences and he has been privately sharing some wonderful integrated phylogenetic analyses covering the entire Ebola outbreak. However, until recently the sharing has been limited by access to public data. At a recent conference at the Institut Pasteur, I met him and his colleague Richard Nehrer and discussed ways to improve sharing. With Trevor Bedford, Richard are the developers of the nextflu website, which aims to track real-time evolution of flu.
I said that we needed this for Ebola, and of course they had already thought of this and had started building something. I said that we would contribute our nanopore sequencing dataset to this project in real-time, and those with large datasets to compare also contributed theirs.
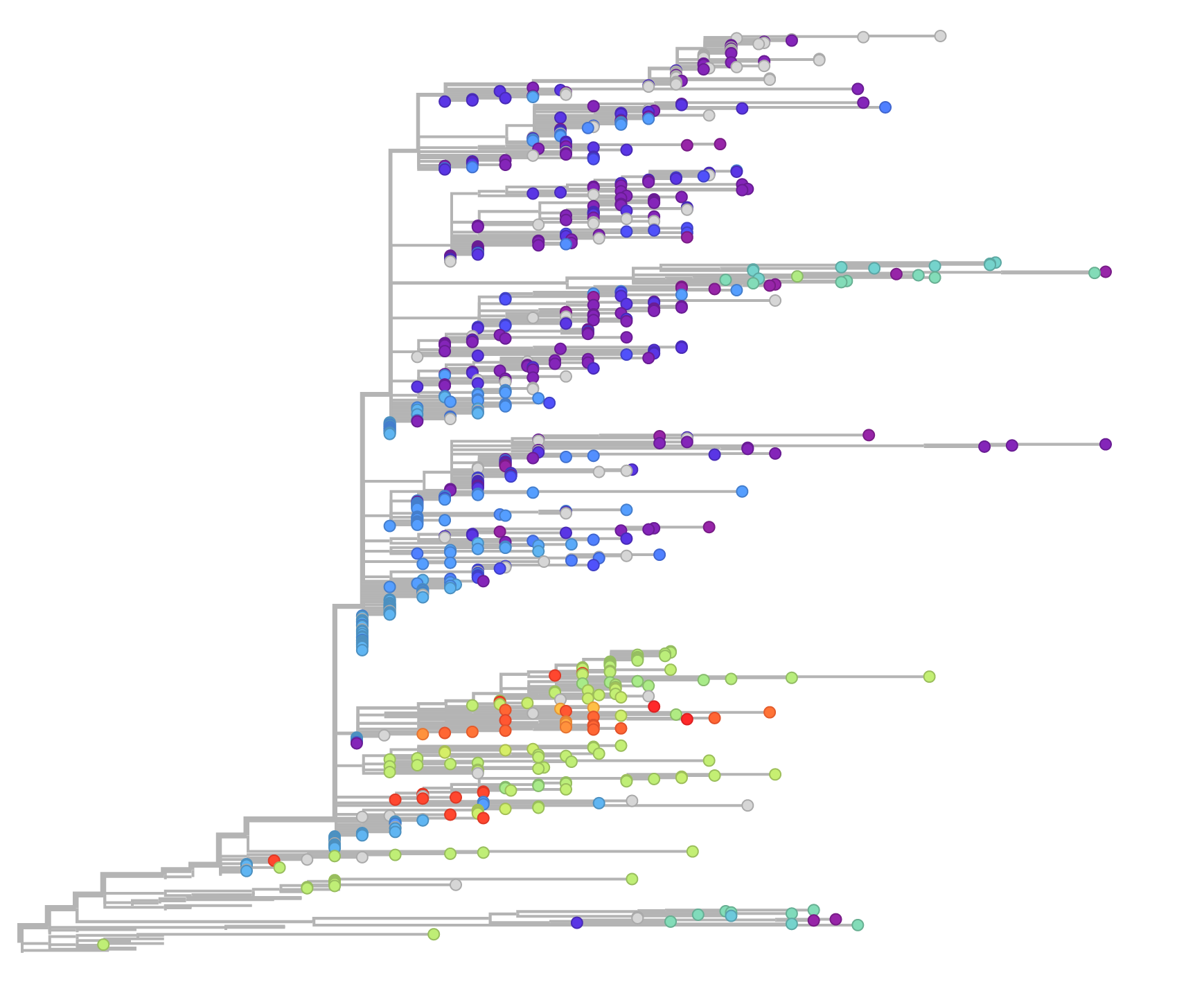
So it is a real thrill to see the website up and running now and available to use at ebola.nextflu.org. On this website you can explore Ebola evolution during this outbreak, using controls to scroll through time, and restricting analysis to particular locations or laboratories. You can also zoom into particular clades, and see frequency distributions of specific mutations.
One thing that was particularly notable with the data integration is that our surveillance data from Guinea, when compared with Ian Goodfellow’s recently produced surveillance data from Sierra Leone is that the two extant Guinean lineages overlap with cases from close to the Guinean border in Sierra Leone. This makes sense, and suggests that cross-country transmission may be frequently occurring.
We will be updating this website with new sequences generated by the EMLab until the end of the outbreak. We have decided that we will leave a one week delay before releasing it for WHO central coordination to see the data, and the data is limited to prefecture level information without more specific locations.